Say Hello to the Scientist Using DNA to Track Plankton Diversity
Ecologist Paula Pappalardo hones strategies for identifying these critical ocean organisms.

On a planet bustling with animal life, identifying every beetle and barnacle you come across can prove a daunting challenge. But researchers are hard at work documenting what kinds of species live where. To do that, they need to identify them correctly, part of a field called taxonomy.
For this month’s “Meet a SI-entist,” we caught up with Paula Pappalardo, an ecologist studying plankton biodiversity at the Smithsonian National Museum of Natural History. She tells us about her work alongside taxonomists, how she’s trying to improve methods to identify species and why plankton communities are key to a healthy ocean.
Most people probably don’t think about plankton on a day-to-day basis. What drew you to invertebrate zoology and these organisms in particular?
I grew up in a small city in Argentina, and I spent a lot of time outdoors, fascinated by insects and spiders. At the same time, my family had a tradition of going on vacation to a small beach town, and I would spend hours walking by the beach looking for tiny seashells. So I think I’ve always liked the little guys.
But I initially went to college to study electronic engineering, not biology. At the end of high school, the career counselor thought that my love of animals looked more like a hobby, and that my skill in math and physics gave me a more logical career path. But during my third year of college, I started a degree in biology. I spent hours reading, and I learned to dive off the coast of Patagonia. I actually did my final thesis at a research institute there, studying clam populations. And I thought, “I want to do this for a living.”

What questions guide your research at the museum?
Most of the astonishing diversity of marine life is hidden in plain sight: tiny and often transparent organisms that drift in the ocean as plankton. I’m interested in their distribution, or why they are where they are. Why are some species here and not there in the ocean, why are some places hotspots of biodiversity and how does the environment affect the distributions of species?
I’m also interested in the methods we use to do research, and if we can optimize those methods for specific questions or for the data that’s available. Right now, I am investigating the best strategies to get an accurate list of species using DNA-based methods.
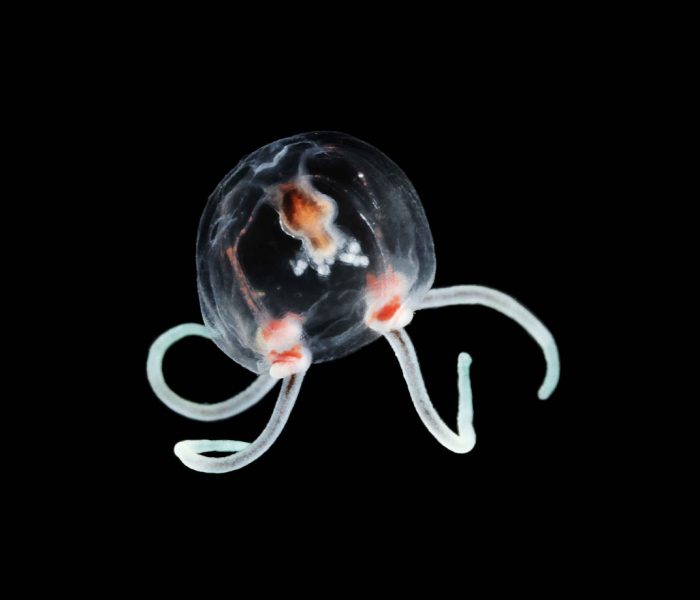
DNA barcoding and metabarcoding are hot topics right now. What are they, and how do they help scientists identify species?
DNA barcoding uses a short portion of DNA from a specific animal to identify it. It’s similar to using a barcode in a grocery store, where you can scan it to identify the specific item. This can be very useful if you’ve collected a fragile organism and it gets damaged, so you’re not able to discern which animal it is, or if you have larvae or juveniles that don’t look similar to the adults.
With metabarcoding, you can do a similar thing, but for a whole community. You could collect the organisms themselves, with something like a plankton net or insect trap. You could also sample DNA from the environment, from things like seawater, sediment or animal feces. This is really useful for organisms that are rare or elusive. They still leave traces behind, and without disturbing the animals, you can find the traces, extract DNA sequences and match them to a DNA reference library to generate a list of organisms present in the sample.
There are tons of decisions a researcher needs to make to generate that final list of species using barcoding and metabarcoding, like which reference library of genetic sequences to use or when you can trust a species assignment. I want to generate specific recommendations that can guide researchers on what strategies work best to identify species in different taxonomic groups.
A lot of your research involves data from the Smithsonian’s StreamCode project a few years ago. Can you tell me a bit about that effort?
The StreamCode project gathered a large group of experts in marine invertebrate taxonomy to collect and identify plankton samples in the Gulf Stream. The Gulf Stream is a very important current that transports organisms and nutrients from the Gulf of Mexico and along the east coast of the U.S. towards Europe. The goal was to describe the diversity of zooplankton in the region and assess how well DNA-based methods measured biodiversity.
The taxonomic experts created a database of DNA sequences from plankton in the Gulf Stream that we then used during the metabarcoding process to identify the organisms in a plankton sample. We found that database much more effective than other public DNA libraries to identify rare and understudied organisms. Our work showed that despite the rapid growth in DNA barcoding and metabarcoding, the results we get with those methods are very dependent on the database we compare them to. The results will be much closer to the real diversity of the water sample if taxonomic experts start by creating a database from the same location.

What does your day-to-day work look like?
I have done fieldwork and lab work for past research, but with the increase in data availability, you can answer large-scale questions about species distributions using computers. I spend multiple hours a day writing code that helps me clean, analyze and visualize data. Sometimes I have to do a task that is too much for my personal computer to handle, so I connect to the Smithsonian High Performance Computing Cluster to do larger computations.
I also clean and double-check a lot of the genetic sequences that our lab generates and put them in public databases so that they’re available to other researchers. Samples can get contaminated with researchers’ DNA in the lab, and the analysis of the sample might say, ‘This is a human.’ It’s certainly not — it’s a snail. So there’s a lot of cleaning of data.
Why is it important to document plankton biodiversity using barcoding, metabarcoding and other strategies?
Diverse planktonic communities are very important for a healthy marine ecosystem. They’re the base of a complex food web that feeds other organisms like fish and whales. They’re also critical for other services we get from the ocean. Tiny phytoplankton produce a lot of the oxygen we breathe, and plankton are very important in the cycle of carbon from the atmosphere to the deep ocean.
Monitoring plankton communities is also critical to understanding and predicting changes that are taking place in our oceans. Climate change is rapidly affecting ocean environments and redistributing marine life. Plankton can be very sensitive to environmental changes, giving us an early-warning system. We need fast and accurate strategies for measuring species distributions in order to understand and manage those changes, and DNA-based methods can help us do that.
We need to keep investing in the groundwork to create comprehensive genetic libraries, and places like the museum will be critical in those efforts.
This interview has been edited for length and clarity.
Meet a SI-entist: The Smithsonian is so much more than its world-renowned exhibits and artifacts. It is a hub of scientific exploration for hundreds of researchers from around the world. Once a month, we’ll introduce you to a Smithsonian Institution scientist (or SI-entist) and the fascinating work they do behind the scenes at the National Museum of Natural History.

Madison Goldberg is an intern in the Smithsonian National Museum of Natural History’s Office of Communications and Public Affairs. She previously spent the summer as a Mass Media Fellow with the American Association for the Advancement of Science, covering energy and the environment for StateImpact Pennsylvania. She holds a BA in Earth and Planetary Sciences with a minor in education from Harvard University.
This post was originally published by the Smithsonian magazine blog, Smithsonian Voices. Copyright 2021 Smithsonian Institution. Reprinted with permission from Smithsonian Enterprises. All rights reserved. Reproduction in any medium is strictly prohibited without permission from Smithsonian Institution.
Posted: 18 March 2022
-
Categories:
Education, Access & Outreach , Feature Stories , Natural History Museum , Science and Nature








